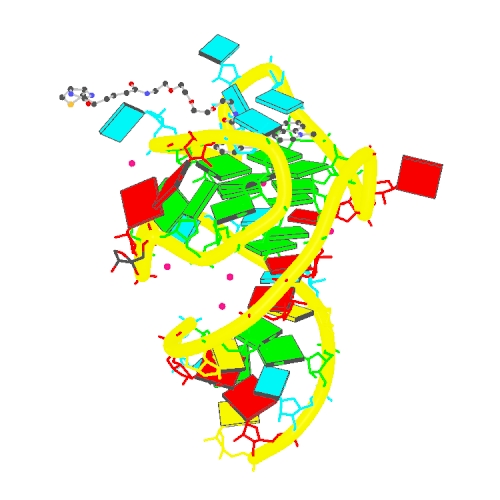
R. J. Trachman 3rd, A. Autour, S. C. Y. Jeng, A. Abdolahzadeh, A. Andreoni, R. Cojocaru, R. Garipov, E. V. Dolgosheina, J. R. Knutson, M. Ryckelynck, P. J. Unrau and A. R. Ferré-DAmaré (2019) Structure and functional reselection of the Mango-III fluorogenic RNA aptamer. Nature Chemical Biology, 15(5): 472-479
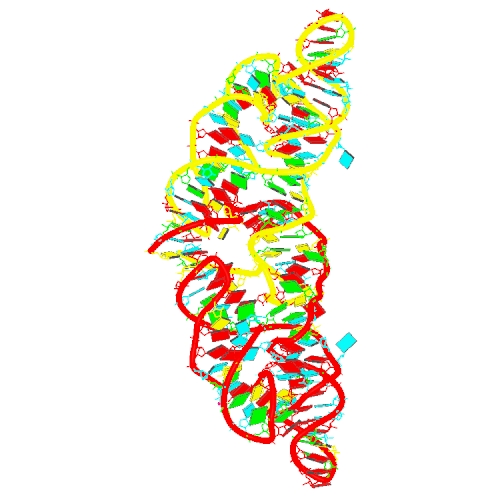
L. Zheng, C. Falschlunger, K. Huang, E. Mairhofer, S. Yuan, J. Wang, D. J. Patel, R. Micura and A. Ren (2019) Hatchet ribozyme structure and implications for cleavage mechanism. Proc Natl Acad Sci U S A., 116(22): 10783-10791
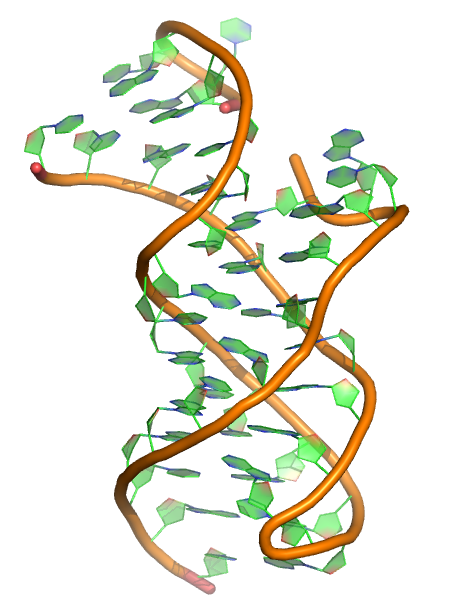
L. Huang, J. Wang, T. J. Wilson and D. M. J. Lilley (2017) Structure of the Guanidine III Riboswitch. Cell Chem Biol, 2017 24(11): 1407-1415.e2.
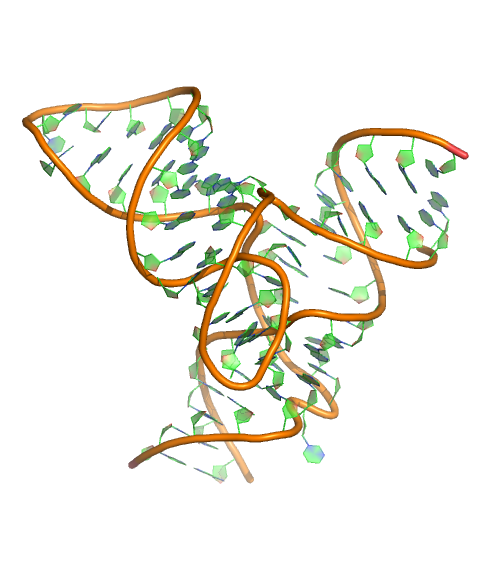
L. Zheng, E. Mairhofer, M. Teplova, Y. Zhang, J. Ma, D. J. Patel, R. Micura and A. Ren (2017) Structure-based insights into self-cleavage by a four-way junctional twister-sister ribozyme. Nature Communications, 8: 1180
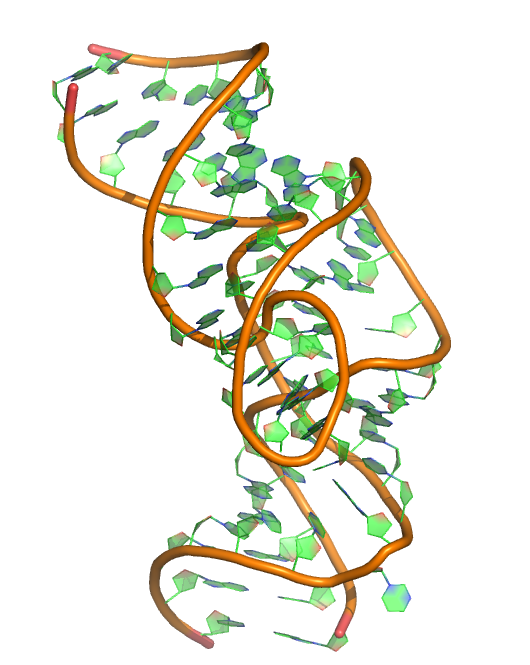
Y. Liu, T. J. Wilson and D. M. J. Lilley (2017) The structure of a nucleolytic ribozyme that employs a catalytic metal ion. Nature Chemical Biology, 13: 508–513